Summary
Actiphage® is a new test that can directly detect the presence of the bacteria that cause bovine tuberculosis and Johne’s disease, so this test is more linked to culture-based methods of detection. Other methods to diagnose these infections rely on detecting different aspects of the animals’ immune response; either tests that detect cell-mediate immune responses, such as those detected by SICCT or gamma interferon, or ELISA tests that detect the production of antibodies in response to infection. Since each of these tests measures different things – and each has their own limitation – trying to compare the performance of these different tests is difficult. In addition, the animal’s immune response to these diseases is complicated, making comparison of test results even more difficult. However, Actiphage can now provide direct evidence of the presence of the bacteria in blood during infection, and the information we are gaining may help us better understand how to best use all of the available tests to tackle these difficult endemic diseases.
Difficulties detecting Mycobacterial infections using standard methods
The gold standard for confirming infection by most types of bacteria is culture, i.e. the organism is grown in the lab from a sample taken from the infected animal. Unfortunately, culturing Mycobacteria (the type of bacteria that cause both Johne’s disease [JD] and bovine TB [bTB]), is very challenging because they grow very, very slowly in the lab and even rapid methods take up to 40 days. Not only is this method very slow, samples have to be treated with harsh chemicals to kill off the fastgrowing organisms which will otherwise swamp the agar plates. However, this treatment also kills some of the Mycobacteria, so there needs to be about 100 cells in a sample to ensure that 1 colony will grow. Hence, although still considered the gold standard method for proving mycobacteria are present in a sample, culturing is not routinely used as a diagnostic.
Instead of culture, most routine tests used to detect both bTB and JD focus on detecting the immune response. In most infections there is a simple relationship between infection and the animal’s response such that there is an increasingly strong response over time. However, Mycobacteria have evolved immune-escaping systems which then allow the establishment of a latent infection and the long, slow development of disease symptoms. It is reported for both JD and bTB that the initial immune response to infection is primarily cellular, and this can be detected using tests such as SICCT and gamma interferon (INF-γ). However, as latent infection is established, the cellular immune responses level off and then start to decline (see Figure 1).
Figure 1: Summary of pattern of immune responses to mycobacterial infections. Based on diagrams produced by Vordermeier et al. (2004) and Rosseels & Huygen (2008)
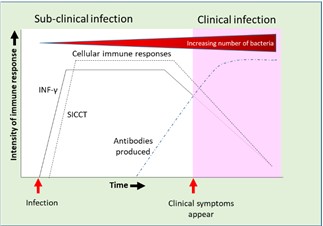
Also associated with the development of latent infection, antibody production is delayed and detectable levels of antibodies are often only produced once symptoms of clinical infection start to become detectable. It seems that the antibody response only really starts to develop once the animal loses control of the latent infection, and then bacterial numbers start to increase and clinical symptoms and pathological signs of infection become evident.
Actiphage – a new approach
Actiphage is based on a method that was developed and approved as a human TB test 20 years ago which was used to detect the Mycobacteria that cause human TB in sputum samples. The method uses a bacteriophage (a virus that is specific for bacterial cells) to detect the presence of the Mycobacteria. In the original assay format, virus particles were allowed to infect the Mycobacteria present in a sample and this process protected the bacteriophage against chemical inactivation. Thus the presence of Mycobacterial cells was indicated because the viruses survived and resulted in to the formation of plaques (zones of clearing) in a lawn of bacteria. Each plaque indicated the presence of one Mycobacterial cell in the original sample, and so the number of Mycobacteria present could be counted (see Figure 2).
Figure 2: Phage-based methods to detect mycobacteria in blood samples. Mycobacteria in blood are found inside white blood cells (PBMCs). These are purified and then lysed to release the mycobacteria. In the Phage-PCR method, the myocbacteria are detected by the formation of plaques and DNA is extracted from these and PCR used to identify the type of cell detected. In the Actiphage method phage are just used to break open the mycobacterial cells to release DNA so that the cells present can be detected and identified by PCR. Diagrams provided by PBD Biotech Ltd. https://www.pbdbio.com/
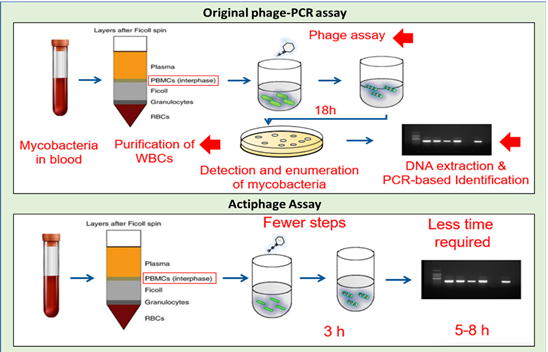
The development that allowed this method to be used to detect animal infections was the discovery that DNA from the Mycobacteria that was being detected was still present in the plaques. This now allowed standard PCR assays, routinely used in labs to identify different types of Mycobacteria, to be used to identify the type of cell that had protected the virus. This method was originally developed to detect Mycobacterium paratuberculosis (MAP) in milk (Stanley et al., 2007), but was then adapted to detect MAP in blood samples (Swift et al., 2013). Then the method was used to try and detect Mycobacterium bovis in blood from SICCT-positive cattle (Swift et al., 2016) and this produced some unexpected results. Although it was well known that MAP could be found in the blood of cattle suffering from JD, it had not been previously shown that M. bovis is circulating in the blood of cattle in the early stages of bTB infection.
However, the original phage-PCR method is quite laborious and can only be carried out in microbiology labs. So, a new version of the method was developed which is now called Actiphage (Figure 2; Swift et al., 2020). This method is simpler, quicker, does not require any agar plates, and was predicted to be more sensitive since there was less opportunity to lose the low levels of DNA during the assay. Indeed, this was found to be the case when comparing results using phage-PCR and Actiphage to test blood samples from animals infected with MAP or M. bovis (Swift et al., 2020).
How do you validate a new test?
Normally new tests are compared with a gold standard method to demonstrate how well it performs. However how do you validate a method that measures something different to the existing tests (in this case SICCT, INF-γ, ELISA) and the gold standard method (i.e. culture) is not very sensitive? If you get more positive results than the gold standard, this can be interpreted as your new test giving false-positive test results.
As Actiphage uses PCR-based detection and identification, surely it would be possible just to compare results with standard PCR test results? Unfortunately, another unusual feature of Mycobacteria comes in to play here because their cell walls contains a thick, waxy layer of mycolic acid that makes them very difficult to break open. Commercial PCR kits that use standard chemical or physical methods to extract DNA from bacteria are therefore not very effective, and the limit of detection of these PCR kits when testing for Mycobacteria is also about 100 cells. The viruses that we use in the Actiphage assay have evolved powerful enzymes that can break the cells open far more efficiently than any chemicals, and so using Actiphage we can routinely detect fewer than 10 cells in a sample using PCR (Swift et al., 2020). So once again, our method is more sensitive than the standard method.
Since the immune response to both JD and bTB is complex, the results for culture (or Actiphage) may or may not agree with other tests, depending on what stage a sample is taken during infection (see Figure 1). We have seen this in our own studies where there has been disagreement between the results gained using commercial JD milk and blood ELISA assays (Swift et al.,2013) and also between JD ELISA and tissue culture results (culture positive animals giving negative blood ELISA results; Swift et al.,2020).
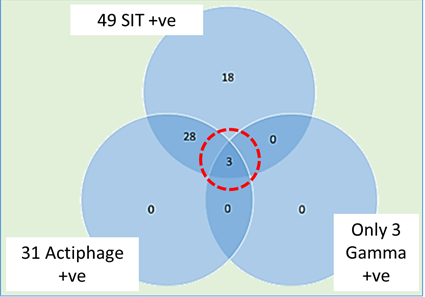
In a study of a herd with a chronic bTB infection, conducted with veterinarian Dick Sibley in Devon, we selected animals on the basis of a positive single intradermal tuberculin (SIT) test result (positive reaction to PPB-B). This version of the skin test is used in many countries as it is considered to be the most sensitive form of the skin test, but it is known to be less specific as cross-reactions in the immune response can occur to other types of Mycobacteria, such as Mycobacterium avium. The animals selected for enhanced testing gave a stronger response to PPB-A than to PPB-B (i.e. were SICCT-negative) and therefore the SIT result was considered to be non-specific. When a set of 49 of these SIT animals were tested with Actiphage, 31 gave a positive test result, confirming that M.bovis was present at detectable levels in their blood (see Figure 3). However, only three animals gave a positive INF-γ test result, and these three animals were also blood ELISA negative – as might be expected as antibody levels are known to rise to detectable levels only after INF-γ responses start to decline (see Figure 1). Again, this points to the fact that we really need to better understand the animals response to infection to understand what test results are telling us and when to use them.
Figure 3: Comparison of Actiphage result with INF-γ.
All this disagreement between test results make the validation process very difficult – which test result do you use as the gold standard? All we can do – and we are actively engaged in this – is to continue to work with farmers and gather data from many different sources until we have a body of information that will satisfy the regulatory bodies, even if on occasions our test does give more positive results than existing methods.
So which test to use, when?
What farmers need to be able to eradicate disease are sensitive, accurate tests to inform their disease management decisions. What we are beginning to understand is that the complexity of the interaction between Mycobacteria and their hosts is not going to make this easy. From all of the data available, we know that when using the most specific forms of these tests – such as SICCT –falsepositive results are unlikely. Our results testing SICCT-positive animals have shown that even NVL animals have detectable levels of M. bovis circulating in their blood (Swift et al., 2016, 2020), and these test results need to be accepted at face value. In JD, using Actiphage to detect the presence of bacteria in the blood indicates that systemic infection has been established, and removes worries about positive faecal culture results that may result from passive transmission or cross-contamination of samples.
Less specific tests than SICCT do run the risk of producing falsepositive results. However, Actiphage only gives a positive test result if the DNA of the bacteria is detected in the blood sample, and we have shown that it can be used in combination with the SIT test to provide confidence that the results are due to M. bovis infections. Tests based on the immune response also have the limitation that they cannot be carried out too close together, leading to long periods when testing cannot be performed. Since Actiphage detects the bacteria rather than an immune response, it can be used at any time and with no need for a gap being left between tests when residual disease can spread to other animals. It can also be used for animals at any age – we have detected both JD and bTB infections in calves. This allows farmers to test young stock and save them the cost of rearing animals that are infected and help the long-term control of disease. Actiphage can also be used to screen animals before they are moved – either within a farm or between farms – to help prevent spread of disease.
However sensitive, all tests are prone to producing false-negative results and test results tend to fluctuate between positive and negative in the early stages of infection before the signal reaches a threshold that can be confidently detected. The complex nature of the biology of Mycobacteria means that antibody responses are slow to develop, and ELISA-based assays are not likely to become consistently positive until the disease is well progressed. The situation is even more complicated for JD and bTB, because in longer, chronic infections the initial cell-mediated immune responses start to decline again, and so infections may then be missed as the disease progresses. Even Actiphage cannot detect infection in its very earliest stages because the levels of mycobacteria in the blood have to get to a sufficient level to allow us to detect it in every blood sample. However, Actiphage now provides a new test option that is not dependent on the vagaries of the immune response and provides definitive evidence that an animal is systemically infected.
Concluding remarks
We are still in the early days of this science, and there is still much to learn about the way that both JD and bTB develop in their hosts. What is becoming clear, however, is there are far more parallels between these two diseases that has often been appreciated. We believe that in addition to providing new information to help disease control programs, Actiphage can also be a new tool that will help us better understand Mycobacterial infections and to pin down exactly which of the available tests is most informative at different stages of infection. Hopefully then we can finally provide a definitive answer to the question of ‘which test to use, when’.
References
Rosseels & Huygen (2008) Vaccination against paratuberculosis. Expert Rev. Vaccines, 7: 817–32.
Vordermeier et al. (2004) The interferon-gamma field trial: background, principles and progress. Vet. Rec. 155: 37–8.
Stanley et al. (2007) Development of a New Rapid Combined Phage and PCR method for the detection and identification of viable Mycobacterium paratuberculosis within 48 h. Appl. Environ. Microbiol. 73: 1851–7.
Swift et al. (2013) Development of a rapid method for the detection of viable Mycobacterium avium subsp. paratuberculosis in blood within 48 h using a phage-based assay. J. Microbiol. Meths. 94: 175–9.
Swift et al. (2016) Evidence of M. tuberculosis Complex bacteraemia in intradermal skin test positive cattle detected using phage-RPA. Virulence, 7: 779–88.
Swift et al. (2020) The development and use of Actiphage to detect viable mycobacteria from bovine tuberculosis and Johne’s disease-infected animals. Microb. Biotechnol. 13: 738–46.